Primary Phagocytic Systems
The principal activity which drives microbivore scaling and design is the process of digestion of organic substances, which alsohas some similarity tothe digestion of food. The microbivore digestive system has four fundamental components -- an array of reversible binding sites to initially bind and trap target microbes, an array of telescoping grapples to manipulate the microbe, once trapped - a morcellation chamber in which the microbe is minced into small, easily digested pieces, and a digestion chamber where the small pieces are chemically digested.
Reversible Microbial Binding Sites
The first function the microbivore must perform is to acquire a pathogen to be digested. A collision between a bacterium of the target species and the nanorobotic device brings their surfaces into intimate contact, allowing reversible binding sites on the microbivore hull to recognize and weakly bind to the bacterium.
Bacterial membranes are quite distinctive, including such obvious markers as the family of outer-membrane trimeric channel proteins called porins in gram-negative bacteria like E. coli and other  surface proteins such as Staphylococcal protein A or endotoxin (lipopolysaccharide or LPS), a variable-size carbohydrate chain that is the major antigen of the outer membrane of gram-negative bacteria. Mycobacteria contain mycolic acid in their cell walls. And only bacteria employ right-handed amino acids in their cellular coats, which helps them resist attack by digestive enzymes in the stomach and by other organisms. Peptidoglycans, the main structural component of bacterial walls, are cross-linked with peptide bridges that contain several unusual nonprotein amino acids and D-enantiomeric forms of Ala, Glu, and Asp. D-alanine is the most abundant D-amino acid found in most peptidoglycans and the only one that is universally incorporated. Macrophages have evolved a variety of plasma membrane receptors that recognize conserved motifs having essential biological roles for pathogens, hence the surface motifs are not subject to high mutation rates; these pathogen receptors on macrophages have been called "pattern recognition receptors" and their targets "pathogen-associated molecular patterns". Genomic differences between virulent and non-pathogenic bacterial strains likely produce phenotypic differences that could enable the biasing of nanorobots towards the detection of the more toxic variants, if necessary.
Additionally, all bacteria of a given species express numerous unique proteins in their outermost coat. Some examples include:single-celled Staphylococcus aureus organism displays binding sites for human vitronectin on its surface; plasmid-specified major outer membrane protein TraTp of Escherichia coli is normally present at the cell surface; Streptococcus pyogenes (strain 6414) has surface binding sites to human collagen; another receptor protein specific to type II collagen (among the dozens of collagen types) are found on the surface of each Staphylococcus aureus (strain Cowan 1) cell. Researchers found that the same bacterial receptor would also specifically respond to synthetic collagen like analogs containing the peptide sequences (Pro-Gly-Pro)n, (Pro-Pro-Gly)10, and (Pro-OH-Pro-Gly)10) If the microbivore must distinguish among ~500 different bacterial species or strains, then each bacterial cell type may be uniquely identified using as few as log2(500) ~ 9 binary antigenic markers. Assuming that nine species-specific bacterial coat ligands are sufficient to uniquely identify an encountered bacterium as belonging to the target species or strain, and that ~104 copies of each of the nine ligands are present on a bacterial surface of area ~10 micron2, then the mean distance between each ligand of the same type is 31.6 nm. A square array of 200 adjacent ligand receptors on the nanorobot surface, with each ligand or receptor active site ~5 nm2 in area (e.g., antibody-antigen complexes typically show contact interfaces of 6-9 nm2, involving 14-21 residues on each side, would on average overlap one such ligand that is resident in a bacterial surface pressed against it. If there are 100 such arrays uniformly distributed over the entire nanorobot surface, then a randomly chosen mutual contact area of only 1% of the nanorobot surface suffices to ensure that there is at least one array overlapping a unique ligand on the bacterial surface during a collision. Of course, the probability of binding, even given mutual contact, is not unity, but perhaps only ~10%. However, this factor is almost completely offset because there are nine equivalent array sets -- one set for each of the nine unique bacterial ligands -- and recognition and binding of any one of the nine unique ligands will suffice to bind the bacterium securely to the nanorobot. Since array members need not be adjacent, the actual physical configuration on the microbivore surface is a bit different. The binding sites are modeled after the narrowband chemical sensor described below.
surface proteins such as Staphylococcal protein A or endotoxin (lipopolysaccharide or LPS), a variable-size carbohydrate chain that is the major antigen of the outer membrane of gram-negative bacteria. Mycobacteria contain mycolic acid in their cell walls. And only bacteria employ right-handed amino acids in their cellular coats, which helps them resist attack by digestive enzymes in the stomach and by other organisms. Peptidoglycans, the main structural component of bacterial walls, are cross-linked with peptide bridges that contain several unusual nonprotein amino acids and D-enantiomeric forms of Ala, Glu, and Asp. D-alanine is the most abundant D-amino acid found in most peptidoglycans and the only one that is universally incorporated. Macrophages have evolved a variety of plasma membrane receptors that recognize conserved motifs having essential biological roles for pathogens, hence the surface motifs are not subject to high mutation rates; these pathogen receptors on macrophages have been called "pattern recognition receptors" and their targets "pathogen-associated molecular patterns". Genomic differences between virulent and non-pathogenic bacterial strains likely produce phenotypic differences that could enable the biasing of nanorobots towards the detection of the more toxic variants, if necessary.
Additionally, all bacteria of a given species express numerous unique proteins in their outermost coat. Some examples include:single-celled Staphylococcus aureus organism displays binding sites for human vitronectin on its surface; plasmid-specified major outer membrane protein TraTp of Escherichia coli is normally present at the cell surface; Streptococcus pyogenes (strain 6414) has surface binding sites to human collagen; another receptor protein specific to type II collagen (among the dozens of collagen types) are found on the surface of each Staphylococcus aureus (strain Cowan 1) cell. Researchers found that the same bacterial receptor would also specifically respond to synthetic collagen like analogs containing the peptide sequences (Pro-Gly-Pro)n, (Pro-Pro-Gly)10, and (Pro-OH-Pro-Gly)10) If the microbivore must distinguish among ~500 different bacterial species or strains, then each bacterial cell type may be uniquely identified using as few as log2(500) ~ 9 binary antigenic markers. Assuming that nine species-specific bacterial coat ligands are sufficient to uniquely identify an encountered bacterium as belonging to the target species or strain, and that ~104 copies of each of the nine ligands are present on a bacterial surface of area ~10 micron2, then the mean distance between each ligand of the same type is 31.6 nm. A square array of 200 adjacent ligand receptors on the nanorobot surface, with each ligand or receptor active site ~5 nm2 in area (e.g., antibody-antigen complexes typically show contact interfaces of 6-9 nm2, involving 14-21 residues on each side, would on average overlap one such ligand that is resident in a bacterial surface pressed against it. If there are 100 such arrays uniformly distributed over the entire nanorobot surface, then a randomly chosen mutual contact area of only 1% of the nanorobot surface suffices to ensure that there is at least one array overlapping a unique ligand on the bacterial surface during a collision. Of course, the probability of binding, even given mutual contact, is not unity, but perhaps only ~10%. However, this factor is almost completely offset because there are nine equivalent array sets -- one set for each of the nine unique bacterial ligands -- and recognition and binding of any one of the nine unique ligands will suffice to bind the bacterium securely to the nanorobot. Since array members need not be adjacent, the actual physical configuration on the microbivore surface is a bit different. The binding sites are modeled after the narrowband chemical sensor described below. 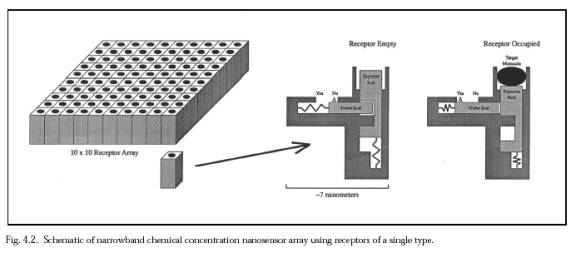 Each 3×3 receptor block consists of nine 7 nm × 7 nm receptor sites, one for each of the nine species-specific bacterial coat ligands. There are 20,000 of these 3×3 receptor blocks distributed uniformly across the microbivore surface. Each 3×3 receptor block measures 21 nm × 21 nm ×10 nm. A single receptor, if bound to a ligand, may provide a binding force of 40-160 pN, probably larger than the largest plausible in sanguo dislodgement force of ~100 pN and thus gripping the bacterium reasonably securely. As an operational procedure, once any one of the nine key ligands has been detected, all of the remaining unoccupied receptors for that ligand in other receptor blocks can be deactivated, and so on until all nine ligands have been individually confirmed -- a combination lock whose completion triggers bacteriocide. Interestingly, during phagocytosis by macrophages most injected particles are recognized by more than one receptor; these receptors are capable of cross-talk and synergy, and phagocytic receptors can both activate and inhibit each other's function.
Microbial binding is energetically favored; if binding energy is ~240 zJ per microbial ligand (1 zeptojoule (zJ) = 10-21 J), then the power requirement for debinding a set of 9 occupied receptors in ~100 microsec is only ~0.02 pW.
Each 3×3 receptor block consists of nine 7 nm × 7 nm receptor sites, one for each of the nine species-specific bacterial coat ligands. There are 20,000 of these 3×3 receptor blocks distributed uniformly across the microbivore surface. Each 3×3 receptor block measures 21 nm × 21 nm ×10 nm. A single receptor, if bound to a ligand, may provide a binding force of 40-160 pN, probably larger than the largest plausible in sanguo dislodgement force of ~100 pN and thus gripping the bacterium reasonably securely. As an operational procedure, once any one of the nine key ligands has been detected, all of the remaining unoccupied receptors for that ligand in other receptor blocks can be deactivated, and so on until all nine ligands have been individually confirmed -- a combination lock whose completion triggers bacteriocide. Interestingly, during phagocytosis by macrophages most injected particles are recognized by more than one receptor; these receptors are capable of cross-talk and synergy, and phagocytic receptors can both activate and inhibit each other's function.
Microbial binding is energetically favored; if binding energy is ~240 zJ per microbial ligand (1 zeptojoule (zJ) = 10-21 J), then the power requirement for debinding a set of 9 occupied receptors in ~100 microsec is only ~0.02 pW.
Telescoping Grapples
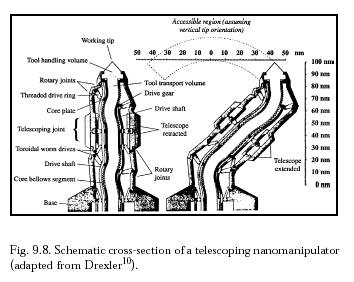
Once the target bacterium has been confirmed and temporarily secured to the microbivore surface at >9 points with a minimum binding force of >360-1440 pN, telescoping robotic grapples emerge from silos in the nanodevice surface to establish secure anchorage to the microbe's plasma membrane or outer coat. Each grapple is mechanically equivalent to the telescoping robotic manipulator arm described by Drexler below  . This manipulator when fully extended is a cylinder 30 nm in diameter and 250 nm in length with a 150-nm diameter work envelope (to the microbivore hull surface), capable of motion up to 1 cm/sec at the tip at a mechanical power cost of ~0.6 pW at moderate load (or ~0.006 pW at 1 mm/sec tip speed), and capable of applying ~1000 pN forces with an elastic deflection of only ~0.1 nm at the tip.
Each telescoping grapple is housed beneath a self-cleaning arising cover mechanism that hides a vertical silo measuring 50 nm in diameter and 300 nm in depth, sufficient to accommodate elevator mechanisms needed to raise the grapple to full extension or to lower it into its fully stowed position. At a 1 mm/sec elevator velocity, the transition requires 0.25 millisec at a Stokes drag power cost (operating in human blood plasma) of 0.0008 pW, or 0.008 pW for 10 grapples maximally extended simultaneously . The elevator mechanism consists of compressed nitrogen gas rotored into or out of the subgrapple chamber volume from a small high-pressure sealed reservoir, a pneumatic piston providing the requisite extension or retraction force. A grapple-distension force of ~100 pN applied for a distance of 250 nm could be provided by 25 atm gas pressure in a minimum subgrapple chamber volume of 104 nm3, involving the importation of ~6000 gas molecules. Removal of these ~6000 gas molecules from a maximum subgrapple chamber volume of 105 nm3 provides a ~1 atm pressure differential and a maximum grapple-retraction force of ~100 pN; cables or other mechanisms may assist in retraction if more force is needed. The aperture of the arising silo cover can be controlled to continuously match the width of the protruding grapple, greatly reducing the intrusion of foreign biomolecules into the silo. Each grapple is terminated with a reversible footpad ~20 nm in diameter. In the case of gram-positive bacteria, a footpad may consist of 100 close-packed lipophilic binding sites targeted to plasma membrane surface lipid molecules, providing a secure 100 pN anchorage between the nanorobot and the bacterium assuming a single-lipid extraction force of ~1 pN. In the case of gram-negative bacteria, a footpad with binding sites for ~3 murein-linked covalently attached transmembrane protein molecules would provide a secure 120-480 pN anchorage, assuming 40-160 pN/molecule and ~9 such molecules per 1000 nm2 of microbial surface. In either case, undesired adhesions with bacterial slime must be avoided. The footpad tool is rotated into, or out of, an exposed position from behind a protective cowling, using countercoiled internal pull cables.
. This manipulator when fully extended is a cylinder 30 nm in diameter and 250 nm in length with a 150-nm diameter work envelope (to the microbivore hull surface), capable of motion up to 1 cm/sec at the tip at a mechanical power cost of ~0.6 pW at moderate load (or ~0.006 pW at 1 mm/sec tip speed), and capable of applying ~1000 pN forces with an elastic deflection of only ~0.1 nm at the tip.
Each telescoping grapple is housed beneath a self-cleaning arising cover mechanism that hides a vertical silo measuring 50 nm in diameter and 300 nm in depth, sufficient to accommodate elevator mechanisms needed to raise the grapple to full extension or to lower it into its fully stowed position. At a 1 mm/sec elevator velocity, the transition requires 0.25 millisec at a Stokes drag power cost (operating in human blood plasma) of 0.0008 pW, or 0.008 pW for 10 grapples maximally extended simultaneously . The elevator mechanism consists of compressed nitrogen gas rotored into or out of the subgrapple chamber volume from a small high-pressure sealed reservoir, a pneumatic piston providing the requisite extension or retraction force. A grapple-distension force of ~100 pN applied for a distance of 250 nm could be provided by 25 atm gas pressure in a minimum subgrapple chamber volume of 104 nm3, involving the importation of ~6000 gas molecules. Removal of these ~6000 gas molecules from a maximum subgrapple chamber volume of 105 nm3 provides a ~1 atm pressure differential and a maximum grapple-retraction force of ~100 pN; cables or other mechanisms may assist in retraction if more force is needed. The aperture of the arising silo cover can be controlled to continuously match the width of the protruding grapple, greatly reducing the intrusion of foreign biomolecules into the silo. Each grapple is terminated with a reversible footpad ~20 nm in diameter. In the case of gram-positive bacteria, a footpad may consist of 100 close-packed lipophilic binding sites targeted to plasma membrane surface lipid molecules, providing a secure 100 pN anchorage between the nanorobot and the bacterium assuming a single-lipid extraction force of ~1 pN. In the case of gram-negative bacteria, a footpad with binding sites for ~3 murein-linked covalently attached transmembrane protein molecules would provide a secure 120-480 pN anchorage, assuming 40-160 pN/molecule and ~9 such molecules per 1000 nm2 of microbial surface. In either case, undesired adhesions with bacterial slime must be avoided. The footpad tool is rotated into, or out of, an exposed position from behind a protective cowling, using countercoiled internal pull cables.
 The tiniest bacterium to be digested may be ~200 nm in diameter , but the smallest virus can be only ~16 nm wide . Since the work envelopes of adjacent grapples picking particles bound to the hull surface extend 150 nm toward each other from either side, the maximum center-to-center intergrapple separation that permits the ciliary transport of 16 nm objects is ~300 nm. This requires 1 grapple per 0.09 micron2 of nanorobot surface, for a total of 277 grapple silos uniformly distributed over the entire 26.885 micron2 microbivore outer hull, excluding the two 1-micron2 port doors. (One or more grapple-containing bridges across the annular exhaust port aperture may be necessary if it is desired to transport targets <200 nm in diameter from the circular DC exhaust port island to the main grapple field of the microbivore, allowing subsequent transport to the ingestion port inlet; such bridges are not included in the present design.) During transport, a bacterium of more typical size such as a 0.4 micron × 2 micron P. aeruginosa bacillus may be supported by up to 9 grapples simultaneously. A somewhat larger E. coli bacterium would be supported by up to 12 grapples. After telescoping grapples are securely anchored to the captive bacterium, the receptor blocks are debonded from the microbial surface, leaving the grapples free to maneuver the pathogen as required. Grapple force sensors inform the onboard computer of the captive microbe's footprint size and orientation. The grapples then execute a ciliary transport protocol in which adjacent manipulators move forward and backward countercyclically, alternately binding and releasing the bacterium, with new grapples along the path ahead emerging from their silos as necessary and unused grapples in the path behind being stowed. Manipulator arrays, ciliary arrays (MEMS), and Intelligent Motion Surfaces are related precursor (and currently available) technologies. Rodlike organisms are first repositioned to align their major axis perpendicular to a great circle plane containing both the device center point and the ingestion port at the front of the device. This keeps the organism traveling over surfaces having the largest possible radius of curvature during transport, thus minimizing any forces necessary to bend the bacterium as it follows the curved microbivore surface. To bend the microbe to the semimajor axis of the microbivore (Rcurve = 1.7 microns) requires F ~ 470 pN, or F ~ 800 pN for the semiminor axis (Rcurve = 1 micron), both of which are substantial bending forces in comparison to the nominal single-grapple anchorage force of 100-500 pN/footpad. Thus it is desirable to bend the bacterium as little as possible during transport. Bending forces may be minimized by adjusting grapple lengths to hold the bacillus farther from the microbivore surface near the endpoints of the footprint, and closer to the microbivore surface near the center of the footprint. Organisms of all shapes are conveyed toward the ingestion port via cyclical ciliary cycling motions. At a transport velocity of 1 mm/sec, a microbe captured at the greatest possible distance from the ingestion port (~3 microns) is moved to the vicinity of the ingestion port in ~3 millisec. The Stokes law energy cost of transporting an E. coli bacterium through blood plasma side-on at 1 mm/sec is 0.01 pW, so transport power is dominated by mechanical losses in the grapples, a total of ~0.06 pW if 10 grapples are operated simultaneously.
Because the ingestion port is slightly recessed into the body of the nanorobot ellipsoid at the equator, the approaching bacterium must be carried around an inlet rim having a considerably smaller radius of curvature than the main body of the microbivore. The inlet rim is essential in this design and provides needed mechanical control from inlet-wall grapples as the microbe is fed into the ingestion port. From simple geometry, if one grapple is fully extended to length L = Lgrap and the adjacent grapple is almost fully retracted to length L ~ 0, then the bacillus can be conveyed around an inlet rim curve of radius Rrim with zero bending if the distance between the adjacent grapples is no more than dmax ~ 2 Rrim sin-1 (Lgrap / 2 Rrim)½ ~ 0.39 microns, taking Lgrap = 250 nm and Rrim ~ 0.25 microns at the inlet rim. This requires at least 1 grapple per dmax2 ~ 0.15 micron2 of nanorobot surface near the ingestion port, comfortably lower in number density than the 0.09 micron2/grapple elsewhere on the hull. Nevertheless, to ensure full control of the transported object near the ingestion port an additional 23 grapple silos are non-uniformly distributed over the 10% of microbivore surface nearest the ingestion port, sufficient to raise the mean number density to 0.05 micron2/grapple in that region. Thus there are a total of 300 grapple silos embedded in the entire microbivore outer hull, excluding the area covered by the two 1-micron2 port doors.
The tiniest bacterium to be digested may be ~200 nm in diameter , but the smallest virus can be only ~16 nm wide . Since the work envelopes of adjacent grapples picking particles bound to the hull surface extend 150 nm toward each other from either side, the maximum center-to-center intergrapple separation that permits the ciliary transport of 16 nm objects is ~300 nm. This requires 1 grapple per 0.09 micron2 of nanorobot surface, for a total of 277 grapple silos uniformly distributed over the entire 26.885 micron2 microbivore outer hull, excluding the two 1-micron2 port doors. (One or more grapple-containing bridges across the annular exhaust port aperture may be necessary if it is desired to transport targets <200 nm in diameter from the circular DC exhaust port island to the main grapple field of the microbivore, allowing subsequent transport to the ingestion port inlet; such bridges are not included in the present design.) During transport, a bacterium of more typical size such as a 0.4 micron × 2 micron P. aeruginosa bacillus may be supported by up to 9 grapples simultaneously. A somewhat larger E. coli bacterium would be supported by up to 12 grapples. After telescoping grapples are securely anchored to the captive bacterium, the receptor blocks are debonded from the microbial surface, leaving the grapples free to maneuver the pathogen as required. Grapple force sensors inform the onboard computer of the captive microbe's footprint size and orientation. The grapples then execute a ciliary transport protocol in which adjacent manipulators move forward and backward countercyclically, alternately binding and releasing the bacterium, with new grapples along the path ahead emerging from their silos as necessary and unused grapples in the path behind being stowed. Manipulator arrays, ciliary arrays (MEMS), and Intelligent Motion Surfaces are related precursor (and currently available) technologies. Rodlike organisms are first repositioned to align their major axis perpendicular to a great circle plane containing both the device center point and the ingestion port at the front of the device. This keeps the organism traveling over surfaces having the largest possible radius of curvature during transport, thus minimizing any forces necessary to bend the bacterium as it follows the curved microbivore surface. To bend the microbe to the semimajor axis of the microbivore (Rcurve = 1.7 microns) requires F ~ 470 pN, or F ~ 800 pN for the semiminor axis (Rcurve = 1 micron), both of which are substantial bending forces in comparison to the nominal single-grapple anchorage force of 100-500 pN/footpad. Thus it is desirable to bend the bacterium as little as possible during transport. Bending forces may be minimized by adjusting grapple lengths to hold the bacillus farther from the microbivore surface near the endpoints of the footprint, and closer to the microbivore surface near the center of the footprint. Organisms of all shapes are conveyed toward the ingestion port via cyclical ciliary cycling motions. At a transport velocity of 1 mm/sec, a microbe captured at the greatest possible distance from the ingestion port (~3 microns) is moved to the vicinity of the ingestion port in ~3 millisec. The Stokes law energy cost of transporting an E. coli bacterium through blood plasma side-on at 1 mm/sec is 0.01 pW, so transport power is dominated by mechanical losses in the grapples, a total of ~0.06 pW if 10 grapples are operated simultaneously.
Because the ingestion port is slightly recessed into the body of the nanorobot ellipsoid at the equator, the approaching bacterium must be carried around an inlet rim having a considerably smaller radius of curvature than the main body of the microbivore. The inlet rim is essential in this design and provides needed mechanical control from inlet-wall grapples as the microbe is fed into the ingestion port. From simple geometry, if one grapple is fully extended to length L = Lgrap and the adjacent grapple is almost fully retracted to length L ~ 0, then the bacillus can be conveyed around an inlet rim curve of radius Rrim with zero bending if the distance between the adjacent grapples is no more than dmax ~ 2 Rrim sin-1 (Lgrap / 2 Rrim)½ ~ 0.39 microns, taking Lgrap = 250 nm and Rrim ~ 0.25 microns at the inlet rim. This requires at least 1 grapple per dmax2 ~ 0.15 micron2 of nanorobot surface near the ingestion port, comfortably lower in number density than the 0.09 micron2/grapple elsewhere on the hull. Nevertheless, to ensure full control of the transported object near the ingestion port an additional 23 grapple silos are non-uniformly distributed over the 10% of microbivore surface nearest the ingestion port, sufficient to raise the mean number density to 0.05 micron2/grapple in that region. Thus there are a total of 300 grapple silos embedded in the entire microbivore outer hull, excluding the area covered by the two 1-micron2 port doors.
 surface proteins such as Staphylococcal protein A or endotoxin (lipopolysaccharide or LPS), a variable-size carbohydrate chain that is the major antigen of the outer membrane of gram-negative bacteria. Mycobacteria contain mycolic acid in their cell walls. And only bacteria employ right-handed amino acids in their cellular coats, which helps them resist attack by digestive enzymes in the stomach and by other organisms. Peptidoglycans, the main structural component of bacterial walls, are cross-linked with peptide bridges that contain several unusual nonprotein amino acids and D-enantiomeric forms of Ala, Glu, and Asp. D-alanine is the most abundant D-amino acid found in most peptidoglycans and the only one that is universally incorporated. Macrophages have evolved a variety of plasma membrane receptors that recognize conserved motifs having essential biological roles for pathogens, hence the surface motifs are not subject to high mutation rates; these pathogen receptors on macrophages have been called "pattern recognition receptors" and their targets "pathogen-associated molecular patterns". Genomic differences between virulent and non-pathogenic bacterial strains likely produce phenotypic differences that could enable the biasing of nanorobots towards the detection of the more toxic variants, if necessary.
surface proteins such as Staphylococcal protein A or endotoxin (lipopolysaccharide or LPS), a variable-size carbohydrate chain that is the major antigen of the outer membrane of gram-negative bacteria. Mycobacteria contain mycolic acid in their cell walls. And only bacteria employ right-handed amino acids in their cellular coats, which helps them resist attack by digestive enzymes in the stomach and by other organisms. Peptidoglycans, the main structural component of bacterial walls, are cross-linked with peptide bridges that contain several unusual nonprotein amino acids and D-enantiomeric forms of Ala, Glu, and Asp. D-alanine is the most abundant D-amino acid found in most peptidoglycans and the only one that is universally incorporated. Macrophages have evolved a variety of plasma membrane receptors that recognize conserved motifs having essential biological roles for pathogens, hence the surface motifs are not subject to high mutation rates; these pathogen receptors on macrophages have been called "pattern recognition receptors" and their targets "pathogen-associated molecular patterns". Genomic differences between virulent and non-pathogenic bacterial strains likely produce phenotypic differences that could enable the biasing of nanorobots towards the detection of the more toxic variants, if necessary.  Each 3×3 receptor block consists of nine 7 nm × 7 nm receptor sites, one for each of the nine species-specific bacterial coat ligands. There are 20,000 of these 3×3 receptor blocks distributed uniformly across the microbivore surface. Each 3×3 receptor block measures 21 nm × 21 nm ×10 nm. A single receptor, if bound to a ligand, may provide a binding force of 40-160 pN, probably larger than the largest plausible in sanguo dislodgement force of ~100 pN and thus gripping the bacterium reasonably securely.
Each 3×3 receptor block consists of nine 7 nm × 7 nm receptor sites, one for each of the nine species-specific bacterial coat ligands. There are 20,000 of these 3×3 receptor blocks distributed uniformly across the microbivore surface. Each 3×3 receptor block measures 21 nm × 21 nm ×10 nm. A single receptor, if bound to a ligand, may provide a binding force of 40-160 pN, probably larger than the largest plausible in sanguo dislodgement force of ~100 pN and thus gripping the bacterium reasonably securely.  . This manipulator when fully extended is a cylinder 30 nm in diameter and 250 nm in length with a 150-nm diameter work envelope (to the microbivore hull surface), capable of motion up to 1 cm/sec at the tip at a mechanical power cost of ~0.6 pW at moderate load (or ~0.006 pW at 1 mm/sec tip speed), and capable of applying ~1000 pN forces with an elastic deflection of only ~0.1 nm at the tip.
. This manipulator when fully extended is a cylinder 30 nm in diameter and 250 nm in length with a 150-nm diameter work envelope (to the microbivore hull surface), capable of motion up to 1 cm/sec at the tip at a mechanical power cost of ~0.6 pW at moderate load (or ~0.006 pW at 1 mm/sec tip speed), and capable of applying ~1000 pN forces with an elastic deflection of only ~0.1 nm at the tip.  The tiniest bacterium to be digested may be ~200 nm in diameter , but the smallest virus can be only ~16 nm wide . Since the work envelopes of adjacent grapples picking particles bound to the hull surface extend 150 nm toward each other from either side, the maximum center-to-center intergrapple separation that permits the ciliary transport of 16 nm objects is ~300 nm. This requires 1 grapple per 0.09 micron2 of nanorobot surface, for a total of 277 grapple silos uniformly distributed over the entire 26.885 micron2 microbivore outer hull, excluding the two 1-micron2 port doors. (One or more grapple-containing bridges across the annular exhaust port aperture may be necessary if it is desired to transport targets <200 nm in diameter from the circular DC exhaust port island to the main grapple field of the microbivore, allowing subsequent transport to the ingestion port inlet; such bridges are not included in the present design.) During transport, a bacterium of more typical size such as a 0.4 micron × 2 micron P. aeruginosa bacillus may be supported by up to 9 grapples simultaneously. A somewhat larger E. coli bacterium would be supported by up to 12 grapples.
The tiniest bacterium to be digested may be ~200 nm in diameter , but the smallest virus can be only ~16 nm wide . Since the work envelopes of adjacent grapples picking particles bound to the hull surface extend 150 nm toward each other from either side, the maximum center-to-center intergrapple separation that permits the ciliary transport of 16 nm objects is ~300 nm. This requires 1 grapple per 0.09 micron2 of nanorobot surface, for a total of 277 grapple silos uniformly distributed over the entire 26.885 micron2 microbivore outer hull, excluding the two 1-micron2 port doors. (One or more grapple-containing bridges across the annular exhaust port aperture may be necessary if it is desired to transport targets <200 nm in diameter from the circular DC exhaust port island to the main grapple field of the microbivore, allowing subsequent transport to the ingestion port inlet; such bridges are not included in the present design.) During transport, a bacterium of more typical size such as a 0.4 micron × 2 micron P. aeruginosa bacillus may be supported by up to 9 grapples simultaneously. A somewhat larger E. coli bacterium would be supported by up to 12 grapples.